10. Chemical Physics – Advanced TAMkin examples¶
TODO: add introductory text.
10.1. Intrinsic rate constants of reactions in zeolites¶
Intrinsic rate constants are typically modeled in a two-step approach where one first describes the reaction in a small (e.g. 5T) cluster model of the zeolite. Given the reactant and transition state geometry with the small cluster, one can construct an initial estimate of the corresponding geometries in a larger cluster using Zeobuilder. These structures can be refined with ONIOM or another type of QM/MM computations.
In the example below we study the addition of ethene to an ethyl alkoxide on an acid site in ZSM-5. The product of this reaction is methylcyclopropane. For the forward reaction rate no computations on the product state are required.
10.1.1. Small cluster computation (5T)¶
The reactant and the transition state are depicted in the figures below
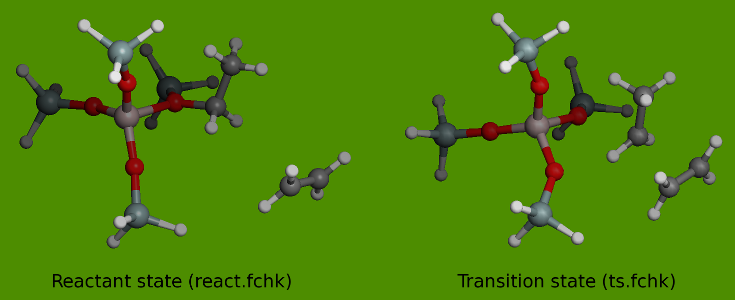
The two checkpoint files in the example are derived from frequency computations with Gaussian03 using the B3LYP/6-31G(d) level of theory at the reactant and transition state.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 |
# Import the tamkin library.
from tamkin import *
# Load the gaussian data:
mol_react = load_molecule_g03fchk("react.fchk")
mol_ts = load_molecule_g03fchk("ts.fchk")
# Perform the normal mode analysis
nma_react = NMA(mol_react, ConstrainExt())
nma_ts = NMA(mol_ts, ConstrainExt(2e-4))
# Construct the two partition functions.
pf_react = PartFun(nma_react, [])
pf_ts = PartFun(nma_ts, [])
# Define a kinetic model for the chemical reaction. These are the mandatory arguments:
# 1) a list of reactant partition functions
# (one for unimolecular, two for bimolecular, ...)
# 2) the transition state partition function
# There is one more optional argument:
# 3) tunneling: a model for the tunelling correction
km = KineticModel([pf_react], pf_ts, tunneling=None)
# Analyze the chemical reaction. These are the arguments:
# 1) A kinetic model
# 2) the starting temperature for the fit
# 3) the final temperature for the fit
# The following argument is optional:
# 4) temp_step: The interval on the temperature grid in Kelvin, 10 is default
ra = ReactionAnalysis(km, 670, 770)
# Make the Arrhenius plot
ra.plot_arrhenius("arrhenius.png")
# Write all results to a file.
ra.write_to_file("reaction.txt")
|
Take note of the following (subtle) choices in the input file:
- Line 8:
- The transition state geometry is not perfectly optimized. This would be a
time-consuming computation and not really required for setting up a draft
model. The gradient_threshold parameter is increased from
1e-4to2e-4. - Line 11-12:
- The translational and rotational contribution can be omitted as these reactions take place on a fixed active site in a zeolite catalyst. Free rotation and translation of the 5T fragment with the guest molecules is not possible. In principle these missing degrees of freedom in the partition function should be replaced by a vibrational coupling of the cluster with the surrounding framework. This is coupling is essentially neglected in a small cluster computation.
10.1.2. Large cluster computation¶
10.2. DIY Kinetic parameter estimation¶
TODO: show how TAMkin can be used to make a powerful freqchk-like
program.
10.3. Many similar reactions with a single script¶
TODO: explain the for-loop.
10.4. Basic kinetics level of theory study¶
TODO
10.5. Smart TAMkin script¶
TODO: explain how to write a smart script with TAMkin that detects, based on a few filename convetions, what type of computation must be carried out. (Specific for Gaussian.)
10.6. Heat of formation¶
TODO: general script for the computation of the heat of formation.
10.7. Pattern matching¶
TODO: how to use the molmod.graphs module to locate a set of frozen
atoms in a series of similar systems with unknown atom order.