5. Auxiliary modules¶
5.1. Timer module¶
-
class
tamkin.timer.Timer¶ Timer object to keep track of the time spent in several parts of TAMKin.
This timer works like a stopwatch. Each time the
sample()method is called the current cpu and wall times are recorded toghether with a labelThe methods
dump()andwrite_to_file()can be used to generate a report.-
dump(f=<open file '<stdout>', mode 'w'>)¶ Dump the logfile with timing information, to screen or to a file stream.
- Optional argument:
- f – the stream to write to. [default=sys.stdout]
-
sample(label)¶ Record the current timings and associate them with the given label
- Argument:
- label – A string describing this point in the code.
-
write_to_file(filename)¶ Write the logfile with timing information to filename.
- Argument:
- filename – the file to write to.
-
5.2. Standard representation of QM or MM data¶
Inheritance diagram
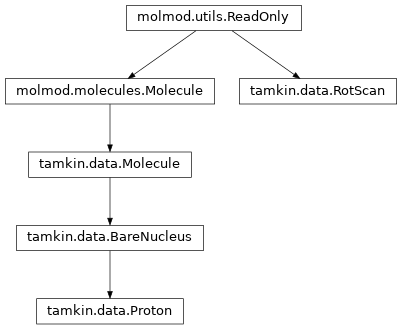
Most IO routines in tamkin.io return instances of the classes
defined here. These objects are just read-only containers for the QM or MM
output with a standardized interface.
-
class
tamkin.data.Molecule(numbers, coordinates, masses, energy, gradient, hessian, multiplicity=None, symmetry_number=None, periodic=False, title=None, graph=None, symbols=None, unit_cell=None, fixed=None)¶ Bases:
molmod.molecules.MoleculeA container for a Hessian computation output from QM or MM codes.
Parameters: - numbers -- The atom numbers (|) –
- coordinates -- the atom coordinates in Bohr (float numpy array (|) – with shape Nx3)
- masses -- The atomic masses in atomic units (float numpy array (|) – with shape N)
- energy -- The molecular energy in Hartree (|) –
- gradient -- The gradient of the energy, i.e. the derivatives (|) – towards Cartesian coordinates, in atomic units (float numpy array with shape Nx3)
- hessian -- The hessian of the energy, i.e. the matrix with (|) – second order derivatives towards Cartesian coordinates, in atomic units (float numpy array with shape 3Nx3N)
- multiplicity -- The spin multiplicity of the electronic system (|) –
- Optional arguments:
symmetry_number– The rotational symmetry number, None when not known or computed [default=None]periodic– True when the system is periodic in three dimensions [default=False]title– The title of the systemgraph– The molecular graph of the systemsymbols– A list with atom symbolsunit_cell– The unit cell vectors for periodic structuresfixed– An array with indices of fixed atoms
-
check_gradient(gradient)¶
-
check_hessian(hessian)¶
-
constrain_ext()¶ Project the global translational and rotational vectors out of the Hessian and the gradient and return a new Molecule instance.
-
coordinates¶ - atomic Cartesian coordinates.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 2.
- Must have shape (?, 3).
- Must have dtype
<type 'numpy.floating'>. - Special conditions: the number of rows must be the same as the length of the array numbers.
Type: *Read-only attribute
-
energy¶ - no documentation available.
The attribute must satisfy the following conditions:
- May not be None.
- Must be an instance of
<type 'float'>
Type: *Read-only attribute
-
external_basis¶ - The basis for small displacements in the external degrees of freedom..
The basis is expressed in mass-weighted Cartesian coordinates. The three translations are along the x, y, and z-axis. The three rotations are about an axis parallel to the x, y, and z-axis through the center of mass. The returned result depends on the periodicity of the system:
- When the system is periodic, only the translation external degrees are included. The result is an array with shape (3,3N)
- When the system is not periodic, the rotational external degrees are also included. The result is an array with shape (6,3N). The first three rows correspond to translation, the latter three rows correspond to rotation.
Type: *Cached attribute
-
fixed¶ - no documentation available.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 1.
- Must have dtype
<type 'numpy.signedinteger'>.
Type: *Read-only attribute
-
get_external_basis_new(im_threshold=1.0)¶ Create a robust basis for small displacements in the external degrees of freedom.
The basis is expressed in mass-weighted Cartesian coordinates. The three translations are along the x, y, and z-axis. The three rotations are about an axis parallel to the x, y, and z-axis through the center of mass.
The returned result depends on the periodicity of the system and on the parameter im_threshold:
- When the system is periodic, only the translation external degrees are included. The result is an array with shape (3,3N)
- When the system is not periodic, the rotational external
degrees are also included. The result is an array with shape (5,3N)
or (6,3N). The first three rows correspond to translation, the
latter rows correspond to rotation. Rotational modes that have a
angular moment below
im_thresholdare discarded.
-
get_submolecule(selected, energy=None, multiplicity=None, symmetry_number=None, periodic=None, graph=None, title=None, symbols=None, unit_cell=None)¶ Create a submolecule with a selection of atoms
- Argument:
selected– list of atom indices, numbering starts at 0- Optional arguments:
energy– Molecular electronic energymultiplicity– The spin multiplicity of the electronic systemsymmetry_number– The rotational symmetry number. Inherited, None when not known or computed [default=0]periodic– True when the system is periodic in three dimensions [default=False]title– The title of the systemgraph– The molecular graph of the systemsymbols– A list with atom symbolsunit_cell– The unit cell vectors for periodic structures
The function returns a Molecule object consisting of the atoms in the selected list. The numbers, coordinates, masses, gradient, and Hessian are reduced in size. The energy, multiplicity, symmetry_number, periodic and symbols attributes are copies of the original molecule attributes, except if they are explicitly specified as optional arguments, then they are overwritten.
-
gradient¶ - no documentation available.
The attribute must satisfy the following conditions:
- May not be None.
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 2.
- Must have shape (?, 3).
- Must have dtype
<type 'numpy.floating'>. - Special conditions: None.
Type: *Read-only attribute
-
graph¶ - the molecular graph with the atom connectivity.
The attribute must satisfy the following conditions:
- Must be an instance of
<class 'molmod.molecular_graphs.MolecularGraph'> - Special conditions: the atomic numbers must match.
Type: *Read-only attribute
-
hessian¶ - no documentation available.
The attribute must satisfy the following conditions:
- May not be None.
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 2.
- Must have dtype
<type 'numpy.floating'>. - Special conditions: None.
Type: *Read-only attribute
-
masses¶ - the atomic masses.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 1.
- Must have dtype
<type 'numpy.floating'>. - Special conditions: the size must be the same as the length of the array numbers.
Type: *Read-only attribute
-
masses3¶ - An array with the diagonal of the mass matrix in Cartesian coordinates..
Each atom mass is repeated three times. The total length of the array is 3N.
Type: *Cached attribute
-
multiplicity¶ - no documentation available.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'int'>
Type: *Read-only attribute
-
numbers¶ - the atomic numbers.
The attribute must satisfy the following conditions:
- May not be None.
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 1.
- Must have dtype
<type 'numpy.signedinteger'>.
Type: *Read-only attribute
-
periodic¶ - no documentation available.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'bool'>
Type: *Read-only attribute
-
raise_ext(shift=1.0)¶ Raise the eigenvalues of the global translations and rotations to a high value, such that their coupling with the internal vibrations becomes negligible, and they can easily be isolated from the vibrations.
-
classmethod
read_from_file(filename)¶ Construct a Molecule object from a previously saved checkpoint file
Parameters: filename -- the file to load from (|) – Usage:
>>> mol = Molecule.read_from_file("mol.chk")
-
symbols¶ - symbols for the atoms, which can be element names for force-field atom types.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'tuple'>
Type: *Read-only attribute
-
symmetry_number¶ - no documentation available.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'int'>
Type: *Read-only attribute
-
title¶ - a short description of the system.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'str'>
Type: *Read-only attribute
-
unit_cell¶ - description of the periodic boundary conditions.
The attribute must satisfy the following conditions:
- Must be an instance of
<class 'molmod.unit_cells.UnitCell'>
Type: *Read-only attribute
-
write_to_file(filename)¶ Write the molecule to a human-readable checkpoint file.
- Argument:
filename– the file to write to
-
class
tamkin.data.BareNucleus(number, mass=None)¶ Bases:
tamkin.data.MoleculeA molecule object for bare nuclei
- Argument:
number– The atom number- Optional argument:
mass– The mass of the atom in atomic units
-
coordinates¶ - atomic Cartesian coordinates.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 2.
- Must have shape (?, 3).
- Must have dtype
<type 'numpy.floating'>. - Special conditions: the number of rows must be the same as the length of the array numbers.
Type: *Read-only attribute
-
energy¶ - no documentation available.
The attribute must satisfy the following conditions:
- May not be None.
- Must be an instance of
<type 'float'>
Type: *Read-only attribute
-
fixed¶ - no documentation available.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 1.
- Must have dtype
<type 'numpy.signedinteger'>.
Type: *Read-only attribute
-
gradient¶ - no documentation available.
The attribute must satisfy the following conditions:
- May not be None.
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 2.
- Must have shape (?, 3).
- Must have dtype
<type 'numpy.floating'>. - Special conditions: None.
Type: *Read-only attribute
-
graph¶ - the molecular graph with the atom connectivity.
The attribute must satisfy the following conditions:
- Must be an instance of
<class 'molmod.molecular_graphs.MolecularGraph'> - Special conditions: the atomic numbers must match.
Type: *Read-only attribute
-
hessian¶ - no documentation available.
The attribute must satisfy the following conditions:
- May not be None.
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 2.
- Must have dtype
<type 'numpy.floating'>. - Special conditions: None.
Type: *Read-only attribute
-
masses¶ - the atomic masses.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 1.
- Must have dtype
<type 'numpy.floating'>. - Special conditions: the size must be the same as the length of the array numbers.
Type: *Read-only attribute
-
multiplicity¶ - no documentation available.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'int'>
Type: *Read-only attribute
-
numbers¶ - the atomic numbers.
The attribute must satisfy the following conditions:
- May not be None.
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 1.
- Must have dtype
<type 'numpy.signedinteger'>.
Type: *Read-only attribute
-
periodic¶ - no documentation available.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'bool'>
Type: *Read-only attribute
-
symbols¶ - symbols for the atoms, which can be element names for force-field atom types.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'tuple'>
Type: *Read-only attribute
-
symmetry_number¶ - no documentation available.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'int'>
Type: *Read-only attribute
-
class
tamkin.data.Proton(mass=None)¶ Bases:
tamkin.data.BareNucleusA molecule object for a proton (or one of its isotopes)
- Optional argument:
mass– The mass of the proton in atomic units
-
coordinates¶ - atomic Cartesian coordinates.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 2.
- Must have shape (?, 3).
- Must have dtype
<type 'numpy.floating'>. - Special conditions: the number of rows must be the same as the length of the array numbers.
Type: *Read-only attribute
-
energy¶ - no documentation available.
The attribute must satisfy the following conditions:
- May not be None.
- Must be an instance of
<type 'float'>
Type: *Read-only attribute
-
fixed¶ - no documentation available.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 1.
- Must have dtype
<type 'numpy.signedinteger'>.
Type: *Read-only attribute
-
gradient¶ - no documentation available.
The attribute must satisfy the following conditions:
- May not be None.
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 2.
- Must have shape (?, 3).
- Must have dtype
<type 'numpy.floating'>. - Special conditions: None.
Type: *Read-only attribute
-
graph¶ - the molecular graph with the atom connectivity.
The attribute must satisfy the following conditions:
- Must be an instance of
<class 'molmod.molecular_graphs.MolecularGraph'> - Special conditions: the atomic numbers must match.
Type: *Read-only attribute
-
hessian¶ - no documentation available.
The attribute must satisfy the following conditions:
- May not be None.
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 2.
- Must have dtype
<type 'numpy.floating'>. - Special conditions: None.
Type: *Read-only attribute
-
masses¶ - the atomic masses.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 1.
- Must have dtype
<type 'numpy.floating'>. - Special conditions: the size must be the same as the length of the array numbers.
Type: *Read-only attribute
-
multiplicity¶ - no documentation available.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'int'>
Type: *Read-only attribute
-
numbers¶ - the atomic numbers.
The attribute must satisfy the following conditions:
- May not be None.
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 1.
- Must have dtype
<type 'numpy.signedinteger'>.
Type: *Read-only attribute
-
periodic¶ - no documentation available.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'bool'>
Type: *Read-only attribute
-
symbols¶ - symbols for the atoms, which can be element names for force-field atom types.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'tuple'>
Type: *Read-only attribute
-
symmetry_number¶ - no documentation available.
The attribute must satisfy the following conditions:
- Must be an instance of
<type 'int'>
Type: *Read-only attribute
-
class
tamkin.data.RotScan(dihedral, molecule=None, top_indexes=None, potential=None)¶ Bases:
molmod.utils.ReadOnlyA container for rotational scan data
- Arguments
dihedral– the index of the atoms that define the dihedral angle- Optional arguments
molecule– a molecule object. required when top_indexes is not giventop_indexes– a list of atom indexes involved in the rotor. required when molecule is not given or when the top_indexes can not be derived automatically.potential– rotational potential info (if this is a hindered rotor). must be a two-tuple containing the angles and the corresponding energies.
-
check_top_indexes(top_indexes)¶
-
dihedral¶ - no documentation available.
The attribute must satisfy the following conditions:
- May not be None.
- Must be an instance of
<type 'numpy.ndarray'> - Must have dimension 1.
- Must have shape (4).
- Must have dtype
<type 'numpy.signedinteger'>.
Type: *Read-only attribute
-
tamkin.data.translate_pbc(molecule, selected, displ, vectors=None)¶ Translate the structure along the lattice vectors
This method is meant to be used in periodic structures, where periodic boundary conditions apply (pbc).
Parameters: - molecule -- A Molecule instance. (|) –
- selected -- A list of indices of the atoms that will be displaced. (|) –
- displ -- A list of 3 integers (|) – [i0,i1,i2]. The selected atoms will be displaced over i0 lattice distances in the 0-axis direction, similarly for i1 and i2.
- Optional argument:
vectors– The lattice vectors, one in each column. If not specified, the vectors in the unit_cell attribute of the molecule is used.
5.3. Analysis of molecular geometries¶
-
tamkin.geom.transrot_basis(coordinates, rot=True)¶ Constructs 6 vectors which represent global translations/rotations.
- Argument:
- coordinates – The atom coordinates (float numpy array with shape Nx3)
- Optional argument:
- rot – When True the rotations are included [default=True]
The return value is a numpy array with 3*N columns and 6 (
rot==True) or 3 (rot==False) rows. The rows are not mass weighted.
-
tamkin.geom.rank_linearity(coordinates, svd_threshold=1e-05, masses3=None)¶ Test the linearity of the given coordinates
Parameters: coordinates -- The atom coordinates (float numpy array with shape (|) – Nx3) - Optional argument:
- svd_threshold – Defines the sensitivity for deviations from linearity [default=1e-5]masses3 – array with atomic masses (each mass is repeated trice)
Returns the number of external degrees of freedom (dof) and the corresponding basis. The dof can be
- 6 dof if atoms of system are non-collinear
- 5 dof if atoms of system are collinear, e.g. when the subsystem contains only 2 atoms
- 3 dof if system contains just 1 atom
If masses3 is given, the analysis is performed in mass-weighted coordinates and the returned basis is in mass-weighted coordinates. If masses3 is not given everything is done in non-mass-weighted Cartesian coordinates.